
CAIDA members Muhammad Abdul-Mageed and Laks V. S. Lakshmanan have recently had a very exciting article published in Nature Communications. You can find the article here.
Authors:
Xiang Zhang, Tianze Ling, Zhi Jin, Sheng Xu, Zhiqiang Gao, Boyan Sun, Zijie Qiu, Jiaqi Wei, Nanqing Dong, Guangshuai Wang, Guibin Wang, Leyuan Li, Muhammad Abdul-Mageed, Laks V. S. Lakshmanan, Fuchu He, Wanli Ouyang, Cheng Chang & Siqi Sun
PrimeNovo: Redefining Protein Sequencing with Revolutionary Precision
This overview was written by Xiang Zhang

In the field of proteomics, scientists are tasked with solving two core challenges: protein structure prediction and protein sequence determination. The former has seen revolutionary breakthroughs with deep learning models like AlphaFold, which have nearly solved the problem of structure prediction. However, the latter—especially mass spectrometry-based protein sequencing—has yet to witness comparable milestones. To address this gap, we introduce PrimeNovo, a novel non-autoregressive Transformer model specifically designed for protein sequencing. PrimeNovo not only overcomes the limitations of existing methods in accuracy and speed but also demonstrates exceptional performance across various biological applications.
The Importance of Protein Sequencing: Decoding the Blueprint of Life
Proteins are the workhorses of life, involved in nearly every vital biological process, from repairing tissues and regulating metabolism to transmitting signals and protecting against diseases. To study proteins, understanding their sequences is paramount. These sequences—long chains of amino acids—serve as the building blocks of life, akin to letters spelling out words. Mass spectrometry (MS) is the primary tool for determining protein sequences. It works by fragmenting proteins into smaller peptides, measuring their masses, and using these measurements to infer the order of amino acids.
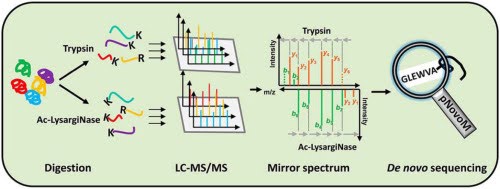
Traditional protein sequencing methods rely heavily on database searches, where experimental MS data is compared against known protein sequences in a database. If a sequence is absent from the database, it cannot be identified. This reliance restricts the discovery of novel proteins, many of which could hold immense significance for scientific research and clinical applications. To overcome this limitation, deep learning-driven de novo sequencing has emerged.
The Current State and Challenges of De Novo Sequencing
De novo sequencing directly infers protein sequences from MS data without relying on databases. This capability has significantly expanded the scope of proteomics. However, most current de novo sequencing models are based on autoregressive frameworks. These models, inspired by successes in natural language processing, predict sequences one amino acid at a time. Despite their innovation, they face several critical limitations.
Autoregressive models generate sequences step-by-step, with each prediction depending on the preceding ones. This approach introduces three main challenges. First, autoregressive models can only utilize information from previously generated parts of the sequence, ignoring the bidirectional dependencies inherent in protein sequences, which limits accuracy. Second, errors made early in the prediction process propagate and compound, leading to deviations in the entire sequence. Third, the step-by-step nature of these models results in slow decoding speeds, making them inefficient for processing large-scale data.
PrimeNovo’s Breakthroughs and Innovations
PrimeNovo is the first non-autoregressive model for protein sequencing, fundamentally transforming the traditional step-by-step generation process. By leveraging the self-attention mechanism in Transformer architecture, PrimeNovo enables each amino acid to consider information from all other positions in the sequence simultaneously. This bidirectional context dramatically improves prediction accuracy.
To address the unique requirements of protein sequencing, PrimeNovo incorporates a Precise Mass Control (PMC) module. Using the total peptide mass provided by MS, the PMC module ensures that the generated sequence adheres strictly to the mass constraints. Unlike traditional models, which rely on iterative search techniques, PMC employs a dynamic programming approach, framing the decoding process as a “knapsack problem.” Each amino acid is treated as an item in the knapsack, with its mass and predicted probability determining its selection. This approach guarantees a globally optimal solution for both sequence and mass accuracy.
Furthermore, PrimeNovo introduces CUDA-optimized parallelized decoding, replacing the sequential processing of autoregressive models. This innovation accelerates prediction speeds by up to 89 times compared to state-of-the-art autoregressive models. This combination of accuracy, speed, and scalability makes PrimeNovo not only suitable for standard protein sequencing tasks but also ideal for high-throughput studies.
Applications in Metaproteomics: Unraveling Complex Protein Communities
Metaproteomics focuses on analyzing the entire set of proteins within complex environmental samples, such as the human gut microbiome or soil ecosystems. These samples often contain a significant proportion of unknown protein sequences, challenging traditional database-dependent methods. PrimeNovo’s non-autoregressive architecture enables direct decoding of novel protein sequences from MS data without relying on existing databases.
In a metaproteomic study involving 17 bacterial strains, PrimeNovo identified 107% more peptide-spectrum matches (PSMs) and 124% more peptides than the leading benchmark model. This dramatic improvement enhances classification accuracy and provides valuable insights into microbial ecological functions. For instance, PrimeNovo can pinpoint unique peptides specific to certain species, shedding light on their roles within complex ecosystems.
Performance in Post-Translational Modification (PTM) Detection
Post-translational modifications (PTMs) such as phosphorylation and acetylation are critical for regulating protein functions and are closely linked to diseases like cancer and metabolic disorders. However, due to their low abundance and diversity, PTMs are difficult to detect using conventional methods.
PrimeNovo’s precise mass control and non-autoregressive decoder excel in identifying low-abundance modified peptides even in complex samples. In a study involving lung adenocarcinoma patients, PrimeNovo successfully distinguished phosphorylated peptides in tumor versus non-tumor tissues with 98% classification accuracy. It also uncovered novel PTMs associated with disease mechanisms, providing a foundation for further research and therapeutic development.
Enhanced Interpretability: Making Deep Learning for Protein Transparent
Deep learning models are often criticized as “black boxes” due to their lack of interpretability. PrimeNovo addresses this by leveraging its self-attention mechanism to reveal which MS peaks contribute most significantly to its predictions. This transparency not only validates model outputs but also offers new biological insights.
For example, in pathogen studies, PrimeNovo can highlight critical peptides influencing its predictions, guiding researchers toward potential vaccine or antibody targets. This feature bridges the gap between computational predictions and experimental validation, empowering researchers with actionable data.
Conclusion: Pioneering a New Era in Proteomics
PrimeNovo is not just a technological innovation; it represents a paradigm shift in protein sequencing and proteomics research. By overcoming the limitations of traditional methods, it delivers unparalleled efficiency and accuracy. Its applications in metaproteomics, PTM detection, and beyond underscore its transformative potential. As scientists continue to explore the mysteries of life, PrimeNovo stands as a vital tool, enabling deeper insights and opening new avenues for fundamental and clinical research.